Raw spectrum import. We currently process only plain ASCII files.
It is recommended that you load raw unprocessed spectra because they contain
plenty of useful statistical information. The format of the file should be like
<m/z value 1>, <intensity value 1>
<m/z value 2>, <intensity value 2>
...
The period (".") is expected as a decimal separator, but you can customise
it before importing the file.
Lucio Andrade and Elias Manolakos, "Signal Background Estimation and Baseline Correction Algorithms for Accurate DNA Sequencing" Journal of VLSI, special issue on Bioinformatics 35:3 pp 229-243 (2003)
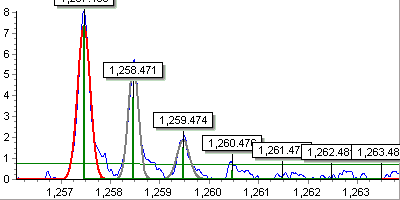
Basic smoothing and local maxima detection. Smoothing here may be important
to improve noise detection for spectra obtained using short aquisition
periods. Such spectra feature coarse quantization artefacts and without smoothing
would perform poorly in noise detection step.
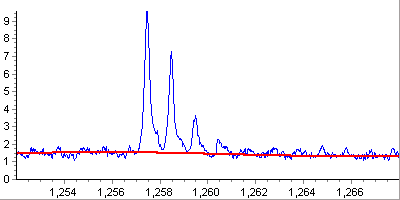
Noise level detection and peak thresholding. This step is crucial in
reducing the number of false peaks resulting from chemical and electronic noise.
Noise level is estimated using Median Absolute Deviation statistic measured
in sliding window and provides satisfactory performance in the wide range
of examined spectra.
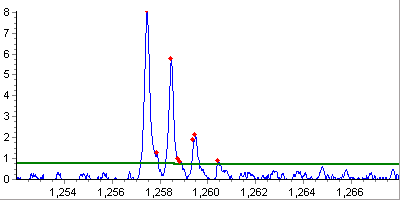
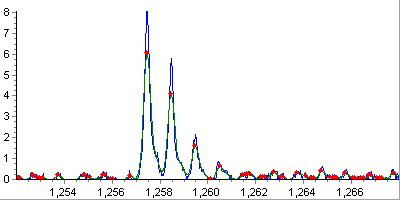
Shape detection. At this step, only the correctly shaped peaks are
retained and their local characteristics like peak width and height are
estimated.
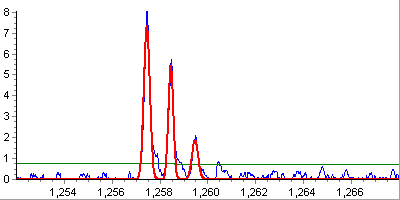
Monoisotopic peaks detection. At this step, algorithm attempts to fit
isotopic distribution at each peak's neighbourhood using various statistical
measures. When successful, the corresponding peak is marked as monoisotopic
and its charge state is estimated.