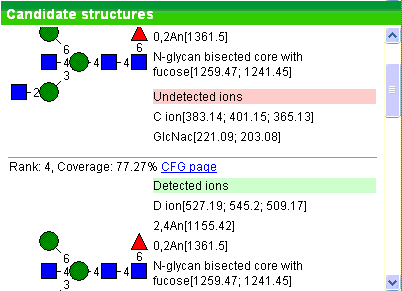
Infer candidate structures using de novo approach. Here algorithm at the first stage
generates structure prototypes as a combination of pre-defined cores and antennae.
Prototypes are ranked based on which diagnostic ions have been detected.
At the second stage, top prototypes yield plausible structures by trying
various linkages. These structures are in turn ranked by number of detected
diagnostic ions and coverage.

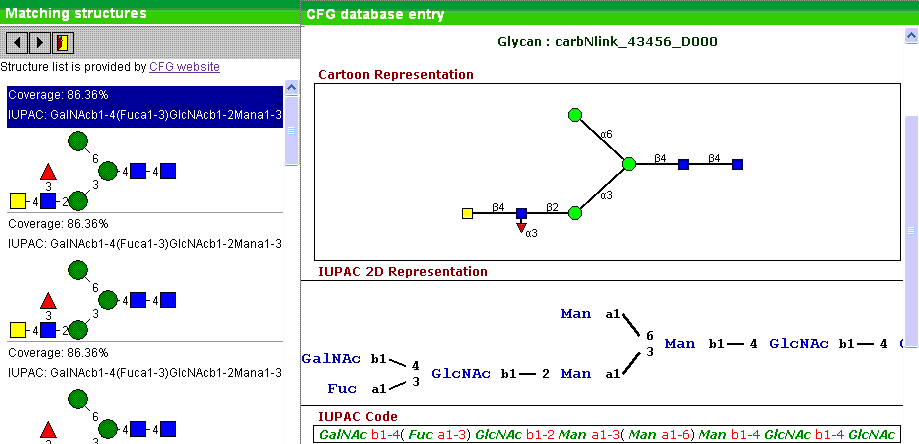
Search open glycan databases. Here we use public database profided by
CDF website. We search the database
by MS1 composition and then rank the structures found by their coverage.
Manually construct and test molecule structures using their own expert knowledge.
Here user will go directly to in silico fragmentation module where they
can simulate and examine structure fragmentation.